The Two Faces of p53
When healthy, p53 senses damaged DNA and is able to make a crucial decision based on its assessment of the extent of damage: to activate the DNA repair machinery and restore the cell back to its healthy state—or to direct it toward cell death, essentially removing it from the system. But the p53 protein is also encoded by a gene—it can be exposed to DNA damage. When this gene mutates, the cell loses a powerful ally. It gets even worse: the mutant p53 protein that is generated in such cells not only loses its genomic guardian function, but it also now becomes capable of facilitating cancer development and progression.
In nature, the non-mutated p53 protein can shift between wild type and mutant-like states. However, when the p53 gene is mutated, the p53 protein becomes permanently locked in a mutant state. We are here to unlock the mutant p53 protein and restore the balance by reinstating its wild type conformation.
Mutant to wild type using a peptide-based strategy
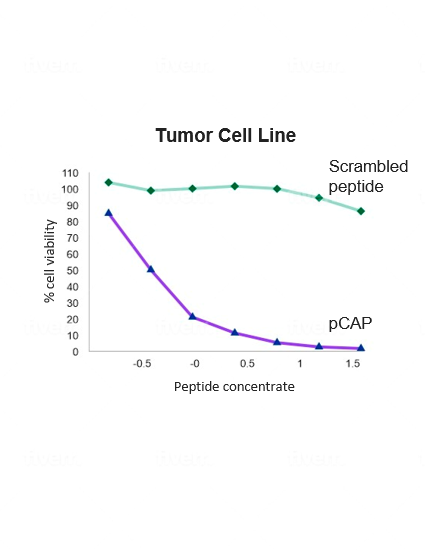
Quintrigen has identified a family of small peptides that can reeducate mutant p53 to do its job. In pre-clinical models, these peptides have been shown to reactivate a wide variety of different mutant p53 proteins by refolding the mutant p53 to its natural native conformation and restoring its potent wild type functionality, including the capability to induce cancer cell death. The treatment is shown to be highly selective against cancer cells, sparing normal cells and tissues.
A peptide-based strategy has many advantages
Restoring a dysfunctional protein to a functional, non-mutated state is a complex undertaking. We believe that our approach is promising and has the potential to treat more than 50% of all human cancers.

Broad and mutation nonspecific +
Our solution reactivates a whole range of p53 mutants, making it applicable to an extensive patient population.

Tumor targeted and highly specific +
Our peptides specifically target mutant p53, which isn’t present in healthy cells.

Promising safety profile +
Because our treatment is tumor-targeted, it leaves healthy cells unharmed, and offers a very promising safety profile.
A structured approach to identify our lead peptide
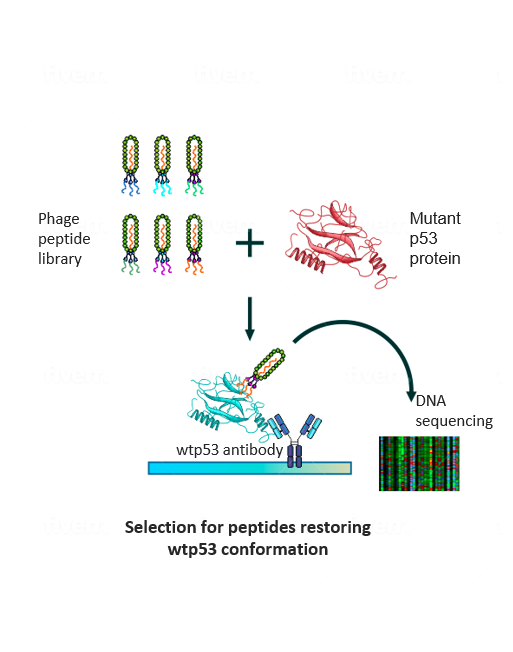
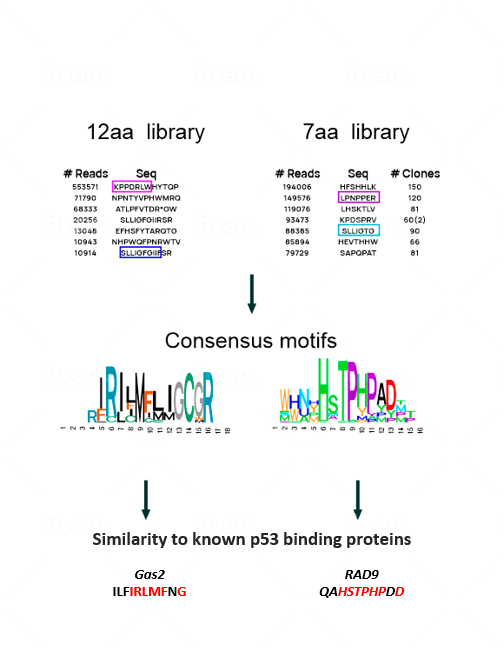
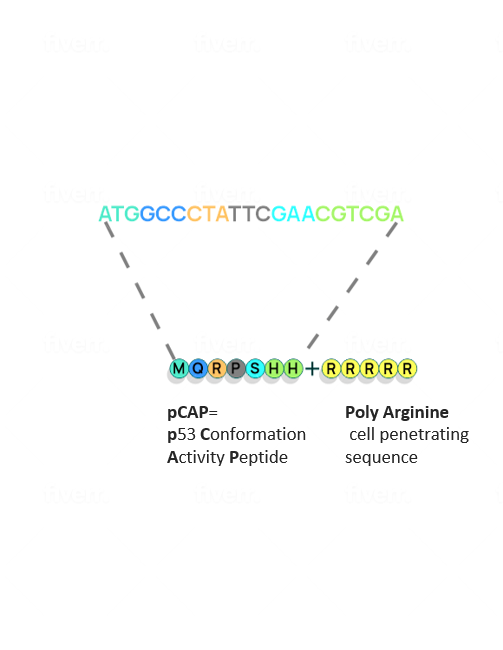
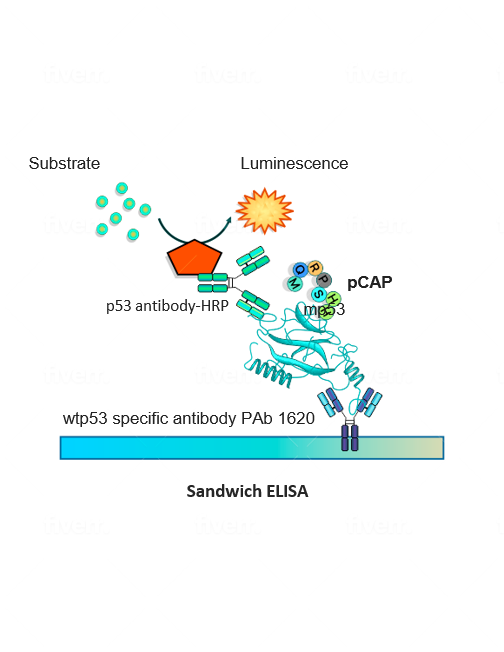
To make the undruggable druggable and return mutated p53 to its rightful place—from bad agent back to trustworthy guardian—new strategies are needed. We have examined libraries of small protein fragments called peptides, which have the potential to enter cells and bind to p53, and have identified those that might teach the mutant protein to regain its original biochemical and biological functions. Finding our target peptide consisted of five main steps:
Publications

Cancer therapeutic approach based on conformational stabilization of mutant p53 protein by small peptides
Cancer therapeutic approach based on conformational stabilization of mutant p53...
Read